 |
Multivariate Pattern Analysis in Python |
 |
Multivariate Pattern Analysis in Python |
If you only have five minutes to decide whether you want to use PyMVPA, take the first minute to look at the following example of a cross-validation procedure on an fMRI dataset (the full source code!). It is not heavily commented, but should simply give you an idea how PyMVPA feels like.
First import the whole PyMVPA module:
>>> from mvpa.suite import *
Now, load the dataset from a NIfTI file. An additional 2-column textfile has the label and associated experimental run of each volume in the dataset (one volume per line). Finally, a mask is loaded to exclude non-brain voxels.
>>> attr = SampleAttributes(os.path.join(pymvpa_dataroot, 'attributes.txt'))
>>> dataset = NiftiDataset(
... samples=os.path.join(pymvpa_dataroot, 'bold.nii.gz'),
... labels=attr.labels,
... chunks=attr.chunks,
... mask=os.path.join(pymvpa_dataroot, 'mask.nii.gz'))
Perform linear detrending and afterwards zscore the timeseries of each voxel using the mean and standard deviation determined from rest volumes (all done for each experimental run individually).
>>> detrend(dataset, perchunk=True, model='linear')
>>> zscore(dataset, perchunk=True, baselinelabels=[0],
... targetdtype='float32')
Select a subset of two stimulation conditions from the whole dataset.
>>> dataset = dataset['labels', [1,2]]
Finally, setup the cross-validation procedure using an odd-even split of the dataset and a SMLR classifier – and run it.
>>> cv = CrossValidatedTransferError(
... TransferError(SMLR()),
... OddEvenSplitter())
>>> error = cv(dataset)
Done. The mean error of classifier predictions on the test dataset across dataset splits is stored in error.
If you think that is a good start, take the remaining four minutes to take a look at the examples shipped in the source distribution of PyMVPA (doc/examples/; some of them are also listed in Full Examples section of this manual). The examples provide a coarse overview of a substantial portion of the functionality provided by PyMVPA, ranging from basic classifier usage, over more sophisticated analysis strategies to simple visualization demos.
All examples are executable scripts that are meant to be run from to toplevel directory of the extracted source tarball, e.g.:
$ doc/examples/start_easy.py
which would run the example shown in the first part of this section.
However, once you found something interesting in the examples you should consider skipping through this manual, as it contains a lot of information that is complementary to the API reference and the examples.
And now for the details ...
The PyMVPA package consists of three major parts: Data handling, Classifiers and various algorithms and measures that operate on datasets and classifiers. In the following sections the basic concept of all three parts will be described and examples using certain parts of the PyMVPA package will be given.
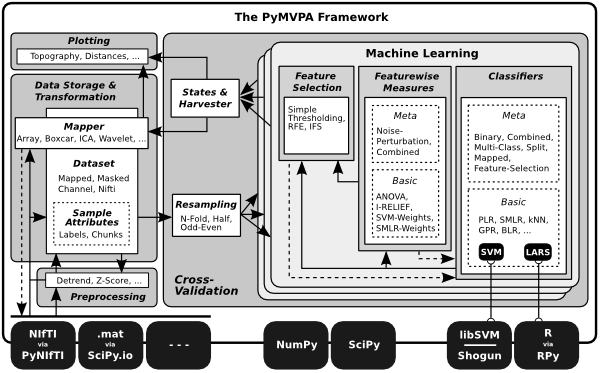
The manual does not cover all bits and pieces of PyMVPA. Detailed information about the module layout and additional documentation about all included functionality is available from the Module Reference – or the API Reference if you are interested in a more technical document. The main purpose of the manual is to give an idea how the individual parts of PyMVPA can be combined to perform complex analyses – easily.