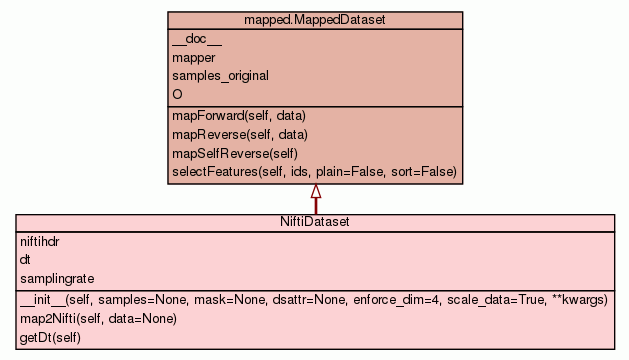
__init__(self,
samples=None,
mask=None,
dsattr=None,
enforce_dim=4,
scale_data=True,
**kwargs)
(Constructor)
| source code
|
If samples and mapper arguments are not None the mapper is
used to forward-map the samples array and the result is passed
to the Dataset constructor.
- Parameters:
samples, str, |, NiftiImage - Filename of a NIfTI image or a NiftiImage instance.mask, str, |, NiftiImage, |, ndarray - Filename of a NIfTI image or a NiftiImage instance or an ndarray
of appropriate shape.enforce_dim (int or None) - If not None, it is the dimensionality of the data to be enforced,
commonly 4D for the data, and 3D for the mask in case of fMRI.scale_data (bool) - NIfTI header specifies scl_slope and scl_inter for scaling and
offsetting the data. By default those will get applied to the data
(change in behavior post 0.4.6). - Overrides:
mapped.MappedDataset.__init__
|