 |
Multivariate Pattern Analysis in Python |
 |
Multivariate Pattern Analysis in Python |
Example showing some possibilities of data exploration (i.e. to ‘smell’ data).
import numpy as N
import pylab as P
import os
from mvpa import pymvpa_dataroot
from mvpa.misc.plot import plotFeatureHist, plotSamplesDistance
from mvpa import cfg
from mvpa.datasets.nifti import NiftiDataset
from mvpa.misc.io import SampleAttributes
from mvpa.datasets.miscfx import zscore, detrend
# load example fmri dataset
attr = SampleAttributes(os.path.join(pymvpa_dataroot, 'attributes.txt'))
ds = NiftiDataset(samples=os.path.join(pymvpa_dataroot, 'bold.nii.gz'),
labels=attr.labels,
chunks=attr.chunks,
mask=os.path.join(pymvpa_dataroot, 'mask.nii.gz'))
# only use the first 5 chunks to save some cpu-cycles
ds = ds.selectSamples(ds.chunks < 5)
# take a look at the distribution of the feature values in all
# sample categories and chunks
plotFeatureHist(ds, perchunk=True, bins=20, normed=True,
xlim=(0, ds.samples.max()))
if cfg.getboolean('examples', 'interactive', True):
P.show()
# next only works with floating point data
ds.setSamplesDType('float')
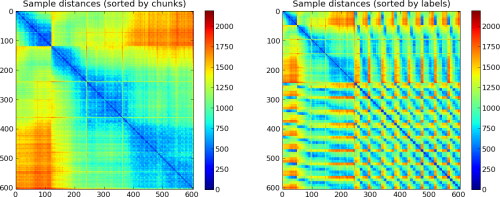
# look at sample similiarity
# Note, the decreasing similarity with increasing temporal distance
# of the samples
P.subplot(121)
plotSamplesDistance(ds, sortbyattr='chunks')
P.title('Sample distances (sorted by chunks)')
# similar distance plot, but now samples sorted by their
# respective labels, i.e. samples with same labels are plotted
# in adjacent columns/rows.
# Note, that the first and largest group corresponds to the
# 'rest' condition in the dataset
P.subplot(122)
plotSamplesDistance(ds, sortbyattr='labels')
P.title('Sample distances (sorted by labels)')
if cfg.getboolean('examples', 'interactive', True):
P.show()
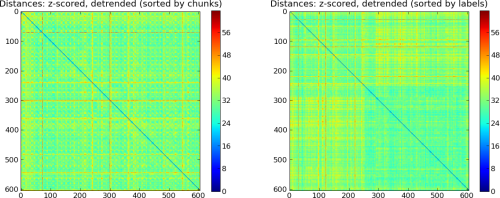
# z-score features individually per chunk
print 'Detrending data'
detrend(ds, perchunk=True, model='regress', polyord=2)
print 'Z-Scoring data'
zscore(ds)
P.subplot(121)
plotSamplesDistance(ds, sortbyattr='chunks')
P.title('Distances: z-scored, detrended (sorted by chunks)')
P.subplot(122)
plotSamplesDistance(ds, sortbyattr='labels')
P.title('Distances: z-scored, detrended (sorted by labels)')
if cfg.getboolean('examples', 'interactive', True):
P.show()
# XXX add some more, maybe show effect of preprocessing
Outputs of the example script. Data prior to preprocessing
Data after minimal preprocessing
See also
The full source code of this example is included in the PyMVPA source distribution (doc/examples/smellit.py).