 |
Multivariate Pattern Analysis in Python |
 |
Multivariate Pattern Analysis in Python |
Demonstrate the usage of a dataset mapper performing data projection onto singular value components within a cross-validation – for any clasifier.
from mvpa.suite import *
if __debug__:
debug.active += ["CROSSC"]
#
# load PyMVPA example dataset
#
attr = SampleAttributes(os.path.join(pymvpa_dataroot, 'attributes.txt'))
dataset = NiftiDataset(samples=os.path.join(pymvpa_dataroot, 'bold.nii.gz'),
labels=attr.labels,
chunks=attr.chunks,
mask=os.path.join(pymvpa_dataroot, 'mask.nii.gz'))
#
# preprocessing
#
# do chunkswise linear detrending on dataset
detrend(dataset, perchunk=True, model='linear')
# only use 'rest', 'cats' and 'scissors' samples from dataset
dataset = dataset.selectSamples(
N.array([ l in [0,4,5] for l in dataset.labels],
dtype='bool'))
# zscore dataset relative to baseline ('rest') mean
zscore(dataset, perchunk=True, baselinelabels=[0], targetdtype='float32')
# remove baseline samples from dataset for final analysis
dataset = dataset.selectSamples(N.array([l != 0 for l in dataset.labels],
dtype='bool'))
print dataset
# Specify the base classifier to be used
# To parametrize the classifier to be used
# Clf = lambda *args:LinearCSVMC(C=-10, *args)
# Just to assign a particular classifier class
Clf = LinearCSVMC
# define some classifiers: a simple one and several classifiers with
# built-in SVDs
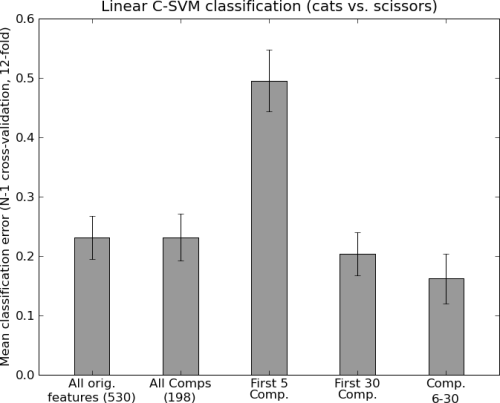
clfs = [('All orig.\nfeatures (%i)' % dataset.nfeatures, Clf()),
('All Comps\n(%i)' % (dataset.nsamples \
- (dataset.nsamples / len(dataset.uniquechunks)),),
MappedClassifier(Clf(), SVDMapper())),
('First 5\nComp.', MappedClassifier(Clf(),
SVDMapper(selector=range(5)))),
('First 30\nComp.', MappedClassifier(Clf(),
SVDMapper(selector=range(30)))),
('Comp.\n6-30', MappedClassifier(Clf(),
SVDMapper(selector=range(5,30))))]
# run and visualize in barplot
results = []
labels = []
for desc, clf in clfs:
print desc
cv = CrossValidatedTransferError(
TransferError(clf),
NFoldSplitter(),
enable_states=['results'])
cv(dataset)
results.append(cv.results)
labels.append(desc)
plotBars(results, labels=labels,
title='Linear C-SVM classification (cats vs. scissors)',
ylabel='Mean classification error (N-1 cross-validation, 12-fold)',
distance=0.5)
if cfg.getboolean('examples', 'interactive', True):
P.show()
Output of the example analysis:
See also
The full source code of this example is included in the PyMVPA source distribution (doc/examples/svdclf.py).